List of my modules...
Module: Watershed
Description:
Computes the morphological watershed on the image named map (on the object pixels), using a seeds object, either an image or a skeleton.
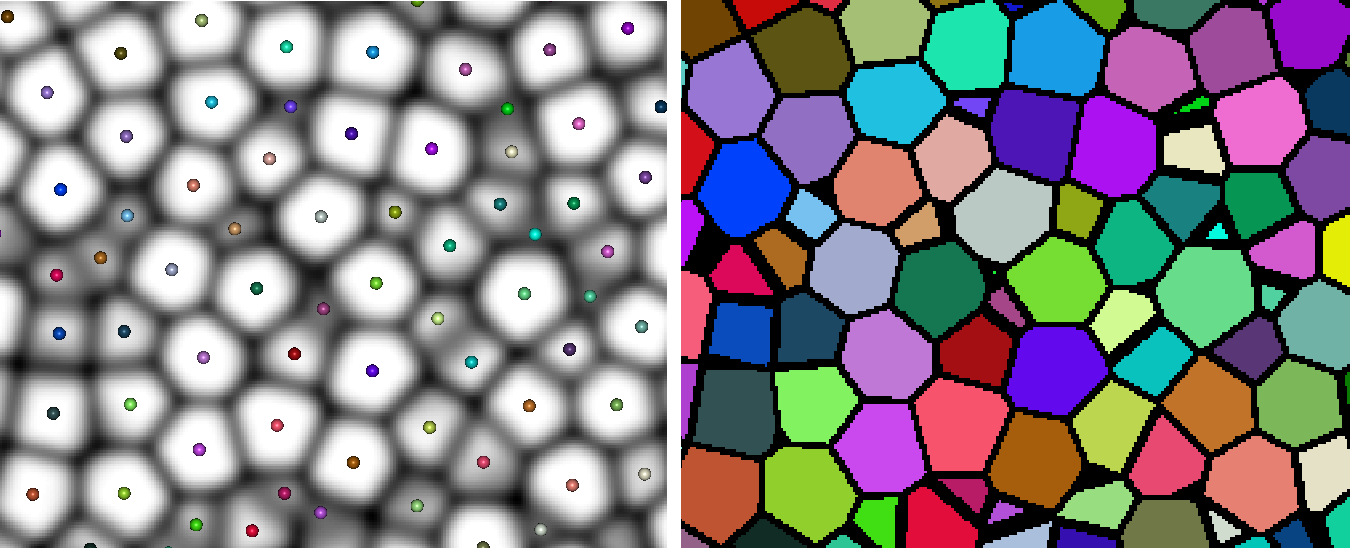
Left: inputs. The set of points represent the seeds, either from the nodes of a HxSkeleton object or the non-zero pixels of a HxUniformScalarField3, colored according to label. The underlying image is the map (inverted in the figure). The right image shows the resulting watershed separation, the black pixels (looks thicker than 1-pixel because it's in 3D and it's a 6-connected watershed).
Connections:
Map
[required]
The input map or topography image, of type HxUniformScalarField3 comes in through this port.
NOTES:
- A threshold is applied to the priority map, this defines the domain in which the watershed will be defined. All pixels not in this domain will be ignored. This is useful when separating a complex object into individual elements. For instance, when you decompose porosity into individual pores, you need not consider the non-porous phase.
- For a seed to identify a region in the output, it must be contained in the thresholded portion of the priority map. Otherwise that label will be discarded in the computation.
Seeds
[required]
The input markers for the regions, either of type HxUniformScalarField3 or of type HxSkeleton.
A seed in a HxUniformScalarField3 is defined as a connected component of pixels that have an identical and non-zero value. It doesn't need not be only one pixel, but can be a whole area of pixels. This can be useful when you already know in what region certain pixels go, in order to speed up the computation and to constrain where the separation will be.
For example, if you want to binarise an image, you can make a rough thresholding of both phases to define a seed image with two seeds (e.g. labelled 1 and 2), with already plenty of pixels in the seeds, and use the inverse of the gradient as a priority image. That way the resulting watershed separation will tend to fit onto the highest gradient regions. A script module named Watershed_Segmentation was made to simplify this process.
NOTES:
- When a skeleton defines the input seeds, a region is defined by all the nodes of the skeleton that have the same label.
- Associated with each node of the skeleton are radii, which define the size of the spheres of influence of each node. When you use the Maximise seeds option, a preprocessing includes all the pixels of these spheres of influence in the seeds before the watershed computation. This is useful when decomposing an object, so that one region isn't gobbled up by a neighbouring one.
Ports:
Threshold

Used to define, along with the comparison operator, the domain in which to apply the watershed.
Comparison

Used to define how the threshold value is used for defining the domain.
Connectivity

Specifies the connectivity, either 6- or 18- or 26-connectivity.
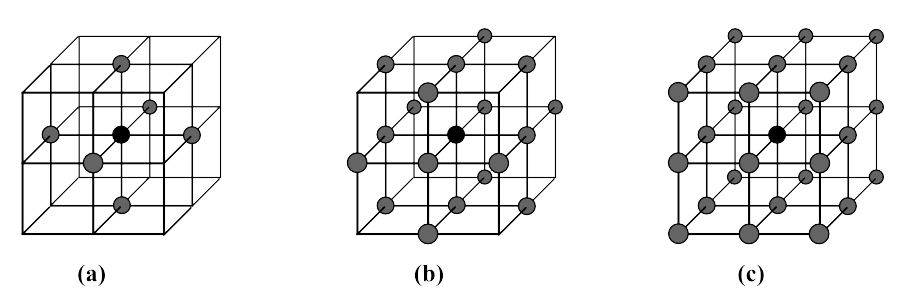
Illustration of the different connectivities in 3D. (a): 6-, (b): 18-, and (c): 26-connectivity (in case you didn't figure it out, n-connectivity means n pixels in the neighbourhood).
Options

The implemented method is a region growth process, where pixels are iteratively assigned to a region, usually until no more pixels are to be assigned, i.e. until convergence. The first option allows you to choose the number of pixels to assign before stopping. The second option is used for a HxSkeleton input for the seed, where each node of the skeleton has an associated value, defining a sphere radius centred on that node. Option maximise seeds assigns, prior to the watershed process, all pixels in the spheres to their respective node labels. The watershed usually creates a pixel-wide separation between regions. The third option allows the pixels of that line to be absorbed by the regions, effectively creating a thin separation.
Iterations

If Convergence was not checked, then this gives the number of pixels to assign to regions before stopping.
Action button

Push the button to start the computation.
Commands:
Additional options can be accessed when typing in the console Watershed COMMAND_NAME. Typing the command again usually reverts back to original settings.
verbose
Displays information on the computation. Retype to hide info.
create
Runs the computation. Returns the name of the output, so it can be used in a script, such as set RESULT [Watershed create].







